LOGISTIC REGRESSION
> setwd("C:/Users/baron/Dropbox/627 Data Analysis/data/My
data")
> Depr = read.csv("depression_data.csv")
#
Another way - reading data directly from the web site:
> Depr = read.csv(url("http://fs2.american.edu/~baron/627/R/depression_data.csv"))
> names(Depr)
[1] "ID"†††††††††††††† "Gender"†††††††††† "Guardian_status"† "Cohesion_score"†
[5] "Depression_score"
"Diagnosis"†
> attach(Depr)
> fix(Data)
> summary(Diagnosis)
†† Min.
1st Qu.† Median††† Mean 3rd Qu.††† Max.†††
NA's
†0.0000† 0.0000† 0.0000†
0.1572† 0.0000† 1.0000†††
2731
#
A lot of missing responses marked as NA. Omit them.
> Depr1 = na.omit(Depr)
> attach(Depr1); dim(Depr1)
[1] 458††
6
#
Now, fit the logistic regression model.
> fit = glm( Diagnosis ~ Gender + Guardian_status
+ Cohesion_score, family = binomial )
> summary(fit)
Coefficients:
†††††††††††††††
Estimate Std. Error z value Pr(>|z|)†††
(Intercept)†††††
1.00832††† 0.50478†† 1.998† 0.04577 *†
GenderMale†††††
-0.68744††† 0.28848† -2.383†
0.01718 *†
Guardian_status -0.74835†††
0.28602† -2.616† 0.00889 **
Cohesion_score† -0.04358†††
0.01046† -4.167 3.09e-05 ***
#
All three variables are significant at 5% level, especially the cohesion score
(connection to community).
#
Cross-validation.
#
How well does our model predict within the training data?
> Prob = fitted.values(fit)
> summary(Prob)
†† Min.
1st Qu.† Median††† Mean 3rd Qu.††† Max.
0.01958 0.07452 0.12990 0.15720 0.21560 0.57710
#
Weíll classify a student as having a depression if the
probability of that exceeds 0.3.
> YesPredict = 1*(Prob > 0.3) ††††† #
For all Prob > 0.3, we let YesPredict
= 1.
††††††††††††††††††††††††††††† ††††† # For all Prob <= 0.3, we let YesPredict
= 0.
#
Then, create a table of true and predicted responses.
> table( Diagnosis, YesPredict )
†††††††† YesPredict
Diagnosis††
0†† 1
0
359† 27
1
†48† 24
# This is not a perfect
result, there are some false positive and false negative diagnoses. Overall, we
correctly predicted (359+24)/458 = 83.6% of cases. The training error rate is only 16.7%. However, among the
students who are really depressed, we correctly
diagnosed only 1/3.
PREDICTION ACCURACY. Training data and test data
# As we know,
prediction error within the training data may be misleading since all responses
were known and used to develop our classification rule. To get a fair estimate
of the correct classification rate, letís
(1)
Split the data into training and test
subsamples;
(2)
Develop the classification rule based on the training
data;
(3)
Use it to classify the test data;
(4)
Cross-tabulate
our prediction with the true classification.
>
n = length(ID)
>
Z = sample(n, n/2)
>
Depr.training = Depr1[ Z, ]
>
Depr.testing = Depr1[ -Z, ]
# Now fit the logistic
model using training data only.
> fit = glm( Diagnosis ~ Gender + Guardian_status
+ Cohesion_score, family = binomial, data = Depr.training )
# Use the obtained rule to classify the test data.
≠≠
>
Prob = predict( fit, data.frame(Depr.testing),
type="response" )
> YesPredict = 1*( Prob > 0.3 )
# Cross-tabulate.
>
attach(Depr.testing)
> table( YesPredict, Diagnosis )
††††††††† Diagnosis
YesPredict†† 0†† 1
†††††††† 0 174† 22
†††††††† †† †††1† 23†
14
# We still classify
80%+ of participants correctly. However, we correctly diagnose only 39% of
students who actually have depression.
Perhaps, gender, parents, and community are not
enough for correct depression diagnostics.
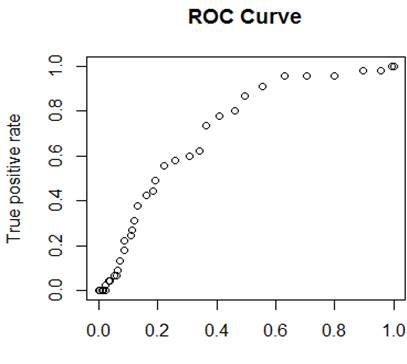
Receiver Operating Characteristic (ROC) Curve
Focus on the true positive rate and the false
positive rate for different thresholds.
True positive rate = P(
predict 1 | true 1 )† = power
False positive rate = P(
predict 1 | true 0 ) = false alarm
>
TPR = rep(0,100); FPR = rep(0,100);
>
for (k in 1:100){
+
fit = glm(Diagnosis ~ Gender
+ Guardian_status + Cohesion_score,
data=Depr1[Z,], family="binomial")
+
Prob = predict( fit, data.frame(Depr1[-Z,]), type="response" )
+
Yhat = (Prob > k/100 )
+
TPR[k] = sum( Yhat==1 &
Diagnosis==1 ) / sum( Diagnosis == 1 )
+
FPR[k] = sum( Yhat==1 &
Diagnosis==0 ) / sum( Diagnosis == 0 )
+
}
>
plot(FPR, TPR, xlab="False
positive rate", ylab="True positive
rate", main="ROC curve")
>
lines(FPR, TPR)
Prediction
Letís predict the diagnosis for some particular
person, a female who lives with both parents, and has an extremely weak
connection with community.
> predict( fit, data.frame(
Gender="Female", Guardian_status=1, Cohesion_score=26 ))
†††††††††
-0.8730466
#
This is the predicted logit. Use the logistic function
to convert it into a probability
> Y0 = predict( fit,
data.frame( Gender="Female", Guardian_status=1, Cohesion_score=26
))
> P0 = exp(Y0)/(1+exp(Y0))
> P0
††††††††
0.2946208
#
This can also be done by the type option.
> predict( fit, data.frame( Gender="Female", Guardian_status=1,
Cohesion_score=26 ), type="response")
†††
0.2946208
#
A 29% chance of developing depression! Suppose she has
an average community connection instead.
> predict( fit, data.frame( Gender="Female", Guardian_status=1,
Cohesion_score=52 ), type="response")
†††††††
0.1185683
#
Only an 11.85% chance now.